Getting started with dxpr: Diagnosis
Hsiang-Ju, Chiu, Yi-Ju Tseng, Chun-Ju, Chen
2022-07-17
Source:vignettes/Eng_Diagnosis.Rmd
Eng_Diagnosis.RmdDescription
The proposed open-source dxpr package is a software tool aimed at expediting an integrated analysis of electronic health records (EHRs). By implementing dxpr package, it is easier to integrate, analyze, and visualize clinical data.
In this part, the instruction of how dxpr package works with diagnosis records is provided.
Development version
install.packages("remotes")
# Install development version from GitHub
remotes::install_github("DHLab-TSENG/dxpr")
library(dxpr)Data Format
dxpr (diagnosis part) is used to pre-process diagnosis codes of EHRs. To execute functions in dxpr, the EHR data input should be a data frame object in R, and contain at least three columns: patient ID, ICD diagnosis codes and date.
Column names or column order of these three columns does not need to necessarily follow a rule. Each required column name will be an argument in functions. Detailed information of required data type of every column and argument of functions can be found in the reference section.
Also, in the R ecosystem, DBI, odbc, and other packages provide access to databases within R. As long as the data is retrieved from databases to a data frame in R, the processes are the same as the following example.
Sample file
A sample rda file is included in dxpr package:
This dataset is a simulated medical dataset of 38 patients with overall
300 records.
head(sampleDxFile)
#> ID ICD Date Version
#> 1: A2 Z992 2020-05-22 10
#> 2: A5 Z992 2020-01-24 10
#> 3: A8 Z992 2015-10-27 10
#> 4: A13 Z992 2020-04-26 10
#> 5: A13 Z992 2025-02-02 10
#> 6: A15 Z992 2023-05-12 10ICD-CM code with two format
dxpr package uses ICD-CM codes as diagnosis standard. There are two
formats of ICD-9 and ICD-10 diagnostic codes, decimal (with a decimal
point separating the code) and short format, respectively. So two tables
of ICD-9-CM and ICD-10-CM are generated to deal with different user
needs: ICD9DxwithTwoFormat and
ICD10DxwithTwoFormat.
ICD-9-CM
# ICD-9-CM_Short
head(ICD9DxwithTwoFormat$Short)
#> [1] "E0000" "E0001" "E0002" "E0008" "E0009" "E0010"
# ICD-9-CM_Decimal
head(ICD9DxwithTwoFormat$Decimal)
#> [1] "E000.0" "E000.1" "E000.2" "E000.8" "E000.9" "E001.0"ICD-10-CM
I.Code Integration
A. ICD diagnostic code format transformation:Short <-> Decimal
dxpr package helps users to standardize the ICD-9 and ICD-10 diagnostic codes into a uniform format before further code grouping. The formats used for different grouping methods are shown as Table 1.
Table 1 Format of code classification methods
| ICD format | |
|---|---|
| Clinical Classifications Software (CCS) | short format |
| Comorbidity | short format |
| Phenome-Wide Association Studies (PheWAS) | decimal format |
Since formats of ICD codes used within a dataset could be different, users can choose a target type (short or decimal) according to the corresponding grouping method.
For example, if a user wants to group data by CCS, then ICD codes should be transformed into short format.
Code standardization for ICD-9 and ICD-10 are executed seperately.
There are two ways to distinguish the version of ICD diagnostic code
(ICD-9/ICD-10) used in data: one is a specific extra column that records
version used (data type in this column should be numeric 9
or 10), the other is a specific date that is the starting
date of using ICD-10 in the dataset. For example, reimbursement claims
with a date required to use ICD-10 codes in the United States and Taiwan
are October 1st, 2015 and January 1st, 2016, respectively.
Warning message
Besides, code standardization functions generate data of diagnosis codes with potential error to help researchers identify the potential coding mistake that may affect the result of following clinical data analysis.
There are two error type:wrong format and
wrong version. The former one means the ICD code does
not exist (maybe because ICD is wrongly coded or with a wrong place of
decimal point). And the latter one means the version is wrong (still use
ICD 9 after icd10usingDate, etc.).
Users can check data after receiving the warning message.
dxpr package also provides an overview of error ICD data by Pareto
chart (function plotICDError).
A-1. Uniform decimal format
The standardization function icdDxShortToDecimal
converts the ICD diagnostic codes to a uniform decimal format, which can
be used for grouping diagnostic code to PheWAS classification.
# Short to decimal
decimal <- icdDxShortToDecimal(dxDataFile = sampleDxFile,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015/10/01")
#> Wrong ICD format: total 9 ICD codes (the number of occurrences is in brackets)
#> c("A0.11 (20)", "E114 (8)", "Z9.90 (6)", "F42 (6)", "001 (5)", "75.52 (4)", "755.2 (3)", "123.45 (3)", "7552 (2)")
#>
#> Wrong ICD version: total 7 ICD codes (the number of occurrences is in brackets)
#> c("V27.0 (18)", "A01.05 (8)", "42761 (7)", "V24.1 (6)", "A0105 (5)", "E03.0 (4)", "650 (4)")
#>
#> Warning: The ICD mentioned above matches to "NA" due to the format or other
#> issues.
#> Warning: "Wrong ICD format" means the ICD has wrong format
#> Warning: "Wrong ICD version" means the ICD classify to wrong ICD version (cause
#> the "icd10usingDate" or other issues)In this example, the starting using date of ICD-10 is “2015/10/01” (format: “YYYY/MM/DD”).
Also, there are 9 ICD codes labeled as “wrong format”, and 7 ICD labeled as “wrong version”.
The results are:
decimal$ICD[6:10]
#> ICD
#> 1: Z99.2
#> 2: 585.5
#> 3: V45.11
#> 4: V56.0
#> 5: 585.3
decimal$Error
#> ICD count IcdVersionInFile WrongType Suggestion
#> 1: A0.11 20 ICD 10 Wrong format
#> 2: V27.0 18 ICD 10 Wrong version
#> 3: E114 8 ICD 10 Wrong format
#> 4: A01.05 8 ICD 9 Wrong version
#> 5: 42761 7 ICD 10 Wrong version
#> 6: Z9.90 6 ICD 10 Wrong format
#> 7: F42 6 ICD 10 Wrong format
#> 8: V24.1 6 ICD 10 Wrong version
#> 9: A0105 5 ICD 9 Wrong version
#> 10: 001 5 ICD 9 Wrong format 0019
#> 11: 75.52 4 ICD 9 Wrong format
#> 12: E03.0 4 ICD 9 Wrong version
#> 13: 650 4 ICD 10 Wrong version
#> 14: 123.45 3 ICD 10 Wrong format
#> 15: 755.2 3 ICD 9 Wrong format 755.29
#> 16: 7552 2 ICD 9 Wrong format 75529decimal$Error shows individual error ICD codes in
descending order.
A-2. Uniform short format
icdDxDecimalToShort function converts the diagnostic
codes to the short format, which can be used for grouping to CCS and
comorbidities classification.
# Decimal to short
short <- icdDxDecimalToShort(dxDataFile = sampleDxFile,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015/10/01")
short$ICD[6:10]
#> ICD
#> 1: Z992
#> 2: 5855
#> 3: V4511
#> 4: V560
#> 5: 5853B. Data integration
Functions in data integration section collapse ICD codes into a smaller number of clinically meaningful categories that are more useful for presenting descriptive statistics than individual ICD diagnostic codes are.
dxpr package supports four strategies to group EHR diagnosis codes, including CCS, PheWAS, comorbidities, and customized defined grouping methods.
The output of code classification contains two data frames.
1) groupedDT
Table 2 groupedDT
| Short/Decimal | ID | ICD | Date | GroupType |
|---|---|---|---|---|
| ICD short/Decimal | patient ID | ICD | Admission date | group of code classification |
The original row order of the data is remained the same in groupedDT,
and only one extra column GroupType is added.
2) summarised_groupedDT
Table 3 summarised_groupedDT
| ID | GroupType | FirstCaseDate | EndCaseDate | Count | Period |
|---|---|---|---|---|---|
| patient ID | group of code classification | first admission date | last admission date | counts of period | record period |
summarised_groupedDT summarised the ICD codes in the same group of the same patient together.
The two outputs can be used in the following functions.
groupedDT can be used to select relevant cases
(function selectCases) and calculate condition era
(function getConditionEra).
summarised_groupedDT can be used to convert the long
format of grouped data into a wide format (function
groupedDataLongToWide) which is fit to other analytical and
plotting packages.
Users can choose the column information of groupType is
“category” or “description” (isDescription =
TRUE or FALSE)
For example, the ccs description is “Tuberculosis” while the category is “1”.
B-1-1. Clinical Classifications Software (CCS)
The CCS classification for ICD-9 and ICD-10 codes is a diagnostic categorization scheme that can employ in many types of projects analyzing data on diagnoses.
Stop updating since 2019.
1) single-level: icdDxToCCS
Both ICD-9-CM and ICD-10-CM code contains 260 single-level CCS categories which can be corresponded with each other.
## ICD to CCS with category description
CCS_description <- icdDxToCCS(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
isDescription = TRUE)
head(CCS_description$groupedDT, 5)
#> Short ID ICD Date CCS_CATEGORY_DESCRIPTION
#> 1: Z992 A2 Z992 2020-05-22 Chronic kidney disease
#> 2: Z992 A5 Z992 2020-01-24 Chronic kidney disease
#> 3: Z992 A8 Z992 2015-10-27 Chronic kidney disease
#> 4: Z992 A13 Z992 2020-04-26 Chronic kidney disease
#> 5: Z992 A13 Z992 2025-02-02 Chronic kidney disease
head(CCS_description$summarised_groupedDT, 5)
#> ID CCS_CATEGORY_DESCRIPTION firstCaseDate endCaseDate count period
#> 1: A0 Chronic kidney disease 2009-07-25 2013-12-20 5 1609 days
#> 2: A1 Chronic kidney disease 2006-11-29 2014-09-24 5 2856 days
#> 3: A10 Chronic kidney disease 2007-11-04 2012-07-30 5 1730 days
#> 4: A11 Chronic kidney disease 2008-03-09 2011-09-03 5 1273 days
#> 5: A12 Chronic kidney disease 2006-05-14 2015-06-29 5 3333 days
## ICD to CCS with category
CCS_category <- icdDxToCCS(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
isDescription = FALSE)
head(CCS_category$groupedDT, 5)
#> Short ID ICD Date CCS_CATEGORY
#> 1: Z992 A2 Z992 2020-05-22 158
#> 2: Z992 A5 Z992 2020-01-24 158
#> 3: Z992 A8 Z992 2015-10-27 158
#> 4: Z992 A13 Z992 2020-04-26 158
#> 5: Z992 A13 Z992 2025-02-02 158
head(CCS_category$summarised_groupedDT, 5)
#> ID CCS_CATEGORY firstCaseDate endCaseDate count period
#> 1: A0 158 2009-07-25 2013-12-20 5 1609 days
#> 2: A1 158 2006-11-29 2014-09-24 5 2856 days
#> 3: A10 158 2007-11-04 2012-07-30 5 1730 days
#> 4: A11 158 2008-03-09 2011-09-03 5 1273 days
#> 5: A12 158 2006-05-14 2015-06-29 5 3333 days2) multi-level: icdDxToCCSLvl
Multi-level CCS in ICD-9-CM has four levels, and multi-level CCS in ICD-10-CM has two levels.
## ICD to CCS multiple level 2 description
CCSlvl_description <- icdDxToCCSLvl(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
CCSLevel = 2,
isDescription = TRUE)
head(CCSlvl_description$groupedDT, 5)
#> Short ID ICD Date CCS_LVL_2_LABEL
#> 1: Z992 A2 Z992 2020-05-22 Diseases of the urinary system
#> 2: Z992 A5 Z992 2020-01-24 Diseases of the urinary system
#> 3: Z992 A8 Z992 2015-10-27 Diseases of the urinary system
#> 4: Z992 A13 Z992 2020-04-26 Diseases of the urinary system
#> 5: Z992 A13 Z992 2025-02-02 Diseases of the urinary system
head(CCSlvl_description$summarised_groupedDT, 5)
#> ID CCS_LVL_2_LABEL firstCaseDate endCaseDate count period
#> 1: A0 Diseases of the urinary system 2009-07-25 2013-12-20 5 1609 days
#> 2: A1 Diseases of the urinary system 2006-11-29 2014-09-24 5 2856 days
#> 3: A10 Diseases of the urinary system 2007-11-04 2012-07-30 5 1730 days
#> 4: A11 Diseases of the urinary system 2008-03-09 2011-09-03 5 1273 days
#> 5: A12 Diseases of the urinary system 2006-05-14 2015-06-29 5 3333 days
## ICD to CCS multiple level 3 category
CCSLvl_category <- icdDxToCCSLvl(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
CCSLevel = 3,
isDescription = FALSE)B-1-2. Clinical Classifications Software Refined (CCSR)
The CCSR classification for ICD-10 codes is a diagnostic categorization scheme that can employ in many types of projects analyzing data on diagnoses.
Unlike CCS, a diagnosis code can be classified into more than one categories in CCSR. However, it is only applicable to ICD-10.
## ICD to CCSR with category description
CCSR_description <- icdDxToCCSR(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
isDescription = TRUE)
head(CCSR_description$groupedDT, 5)
#> Short ID ICD Date CCSR_CATEGORY_DESCRIPTION
#> 1: Z992 A2 Z992 2020-05-22 Other specified status
#> 2: Z992 A5 Z992 2020-01-24 Other specified status
#> 3: Z992 A8 Z992 2015-10-27 Other specified status
#> 4: Z992 A13 Z992 2020-04-26 Other specified status
#> 5: Z992 A13 Z992 2025-02-02 Other specified status
head(CCSR_description$summarised_groupedDT, 5)
#> ID CCSR_CATEGORY_DESCRIPTION firstCaseDate endCaseDate count period
#> 1: A13 Other specified status 2020-04-26 2025-02-02 2 1743 days
#> 2: A15 Other specified status 2023-05-12 2023-05-12 1 0 days
#> 3: A2 Other specified status 2020-05-22 2020-05-22 1 0 days
#> 4: A5 Other specified status 2020-01-24 2020-01-24 1 0 days
#> 5: A8 Other specified status 2015-10-27 2015-10-27 1 0 daysB-2. PheWAS
The dxpr package applied PheWAS, performing a hierarchical grouping of ICD-9 diagnostic codes and ICD-10 diagnostic codes (beta version).
## ICD to PheWAS
PheWAS <- icdDxToPheWAS(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
isDescription = FALSE)
PheWAS$groupedDT[7:11]
#> Decimal ID ICD Date PheCode
#> 1: 585.5 A0 5855 2013-12-20 585.32
#> 2: V45.11 A0 V4511 2012-04-05 585.31
#> 3: V56.0 A0 V560 2010-03-28 585.31
#> 4: 585.3 A0 5853 2010-10-29 585.33
#> 5: 585.6 A0 5856 2009-07-25 585.32
PheWAS$summarised_groupedDT[7:11]
#> ID PheCode firstCaseDate endCaseDate count period
#> 1: A0 585.31 2010-03-28 2012-04-05 2 739 days
#> 2: A0 585.33 2010-10-29 2010-10-29 1 0 days
#> 3: A1 585.32 2006-11-29 2013-04-28 3 2342 days
#> 4: A1 585.33 2014-09-24 2014-09-24 1 0 days
#> 5: A1 585.31 2008-06-25 2008-06-25 1 0 daysB-3. Comorbidities
The dxpr package provides three grouping methods of comorbidity as below:
1) AHRQ
AHRQ comorbidity measure dataset is based on AHRQ Elixhauser Comorbidity Index.
AHRQ <- icdDxToComorbid(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
comorbidMethod = AHRQ)
AHRQ$groupedDT[160:164]
#> Short ID ICD Date Comorbidity
#> 1: M06821 D1 M06821 2016-12-06 Rheumatic
#> 2: K7291 D1 K7291 2024-04-04 Liver
#> 3: O99353 D2 O99353 2023-10-23 NeuroOther
#> 4: F13230 D2 F13230 2022-09-15 Drugs
#> 5: C8397 D2 C8397 2019-10-13 Lymphoma
head(AHRQ$summarised_groupedDT, 5)
#> ID Comorbidity firstCaseDate endCaseDate count period
#> 1: A0 Renal 2009-07-25 2013-12-20 5 1609 days
#> 2: A1 Renal 2006-11-29 2014-09-24 5 2856 days
#> 3: A10 Renal 2007-11-04 2012-07-30 5 1730 days
#> 4: A11 Renal 2008-03-09 2011-09-03 5 1273 days
#> 5: A12 Renal 2006-05-14 2015-06-29 5 3333 days2) Charlson
Charlson comorbidity measure dataset is based on Quan’s translations of the Charlson Comorbidity Index.
Charlson <- icdDxToComorbid(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
comorbidMethod = charlson)
Charlson$groupedDT[160:164]
#> Short ID ICD Date Comorbidity
#> 1: M06821 D1 M06821 2016-12-06 Rheum
#> 2: K7291 D1 K7291 2024-04-04 MSLD
#> 3: O99353 D2 O99353 2023-10-23 <NA>
#> 4: F13230 D2 F13230 2022-09-15 <NA>
#> 5: C8397 D2 C8397 2019-10-13 CANCER
head(Charlson$summarised_groupedDT, 5)
#> ID Comorbidity firstCaseDate endCaseDate count period
#> 1: A0 RD 2009-07-25 2013-12-20 5 1609 days
#> 2: A1 RD 2006-11-29 2014-09-24 5 2856 days
#> 3: A10 RD 2007-11-04 2012-07-30 5 1730 days
#> 4: A11 RD 2008-03-09 2011-09-03 5 1273 days
#> 5: A11 DIAB_C 2015-12-16 2015-12-16 1 0 days3) Elixhauser
The Elixhauser comorbidity software is one in a family of databases and software tools developed as part of the Healthcare Cost and Utilization Project (HCUP).
ELIX <- icdDxToComorbid(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
comorbidMethod = elix)
ELIX$groupedDT[160:164]
#> Short ID ICD Date Comorbidity
#> 1: M06821 D1 M06821 2016-12-06 ARTH
#> 2: K7291 D1 K7291 2024-04-04 LIVER
#> 3: O99353 D2 O99353 2023-10-23 NEURO
#> 4: F13230 D2 F13230 2022-09-15 DRUG
#> 5: C8397 D2 C8397 2019-10-13 LYMPH
head(ELIX$summarised_groupedDT, 5)
#> ID Comorbidity firstCaseDate endCaseDate count period
#> 1: A0 RENLFAIL 2009-07-25 2013-12-20 5 1609 days
#> 2: A1 RENLFAIL 2006-11-29 2014-09-24 5 2856 days
#> 3: A10 RENLFAIL 2007-11-04 2012-07-30 5 1730 days
#> 4: A11 RENLFAIL 2008-03-09 2011-09-03 5 1273 days
#> 5: A12 RENLFAIL 2006-05-14 2015-06-29 5 3333 daysB-4. Customize group method
The dxpr package provided customized grouping functions, in which researches can define the grouping categories; therefore, it is more flexible for grouping ICD diagnostic codes.
For example, researcher can declare a customized grouping table for chronic kidney disease category, and convert an existing dataset into a grouped chronic kidney disease dataset.
There are two functions for customized defined grouping method based on precise and fuzzy grouping method, respectively.
1) Precise method: icdDxToCustom
# CustomGroupingTable
groupingTable <- data.frame(Group = rep("Chronic kidney disease",6),
ICD = c("N181","5853","5854","5855","5856","5859"),
stringsAsFactors = FALSE)
CustomGroup <- icdDxToCustom(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
customGroupingTable = groupingTable)
CustomGroup$groupedDT[10:14]
#> ICD ID Date Group
#> 1: 5853 A0 2010-10-29 Chronic kidney disease
#> 2: 5856 A0 2009-07-25 Chronic kidney disease
#> 3: 5856 A1 2006-11-29 Chronic kidney disease
#> 4: 5855 A1 2012-06-19 Chronic kidney disease
#> 5: 5855 A1 2013-04-28 Chronic kidney disease
head(CustomGroup$summarised_groupedDT, 5)
#> ID Group firstCaseDate endCaseDate count period
#> 1: A0 Chronic kidney disease 2009-07-25 2013-12-20 3 1609 days
#> 2: A1 Chronic kidney disease 2006-11-29 2014-09-24 4 2856 days
#> 3: A10 Chronic kidney disease 2007-11-04 2007-11-04 1 0 days
#> 4: A11 Chronic kidney disease 2008-03-09 2010-02-21 2 714 days
#> 5: A12 Chronic kidney disease 2006-05-14 2011-02-25 3 1748 days2) Fuzzy method: icdDxToCustomGrep
# CustomGroupingTable
grepTable <- data.frame(Group = "Chronic kidney disease",
grepIcd = "^585|^N18",
stringsAsFactors = FALSE)
CustomGrepGroup <- icdDxToCustomGrep(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
customGroupingTable = grepTable)
CustomGrepGroup$groupedDT[10:14]
#> ID ICD Date GrepedGroup
#> 1: A0 5853 2010-10-29 Chronic kidney disease
#> 2: A0 5856 2009-07-25 Chronic kidney disease
#> 3: A1 5856 2006-11-29 Chronic kidney disease
#> 4: A1 5855 2012-06-19 Chronic kidney disease
#> 5: A1 5855 2013-04-28 Chronic kidney disease
head(CustomGrepGroup$summarised_groupedDT, 5)
#> ID GrepedGroup firstCaseDate endCaseDate count period
#> 1: A0 Chronic kidney disease 2009-07-25 2013-12-20 3 1609 days
#> 2: A1 Chronic kidney disease 2006-11-29 2014-09-24 4 2856 days
#> 3: A10 Chronic kidney disease 2007-11-04 2007-11-04 1 0 days
#> 4: A11 Chronic kidney disease 2008-03-09 2010-02-21 2 714 days
#> 5: A12 Chronic kidney disease 2006-05-14 2011-02-25 3 1748 daysII. Data Wrangling
A. Cases selection
The query function selectCases can select the cases
matching defined case conditions (been diagnosed with certain condition
for certain times within a specific duration). User can select cases by
diagnostic categories, such as CCS category, ICD codes, etc.
The output of this function provides the start and end dates of the cases, the number of days between them, and the most common ICD codes used in the case definition.
Case <- selectCases(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
groupDataType = ccslvl2,
icd10usingDate = "2015/10/01",
caseCondition = "Diseases of the urinary system",
caseCount = 1,
caseName = "Selected")
head(Case)
#> ID selectedCase count firstCaseDate endCaseDate period MostCommonICD
#> 1: A3 Selected 5 2008-07-08 2014-02-24 2057 days V420
#> 2: A1 Selected 5 2006-11-29 2014-09-24 2856 days 5855
#> 3: A10 Selected 5 2007-11-04 2012-07-30 1730 days V5631
#> 4: A12 Selected 5 2006-05-14 2015-06-29 3333 days 5859
#> 5: A13 Selected 5 2006-04-29 2025-02-02 6854 days 5855
#> 6: A15 Selected 5 2007-05-25 2023-05-12 5831 days V5631
#> MostCommonICDCount
#> 1: 3
#> 2: 2
#> 3: 2
#> 4: 2
#> 5: 2
#> 6: 2B. Get eligible period of patient records
The function getEligiblePeriod is used for querying the
earliest and latest admission date for each patient.
admissionDate <- getEligiblePeriod(dxDataFile = sampleDxFile,
idColName = ID,
dateColName = Date)
head(admissionDate)
#> ID firstRecordDate endRecordDate
#> 1: D6 2005-10-09 2025-01-05
#> 2: A12 2006-01-12 2022-06-12
#> 3: D1 2006-02-12 2024-04-04
#> 4: A13 2006-04-29 2025-02-02
#> 5: A9 2006-06-30 2023-12-10
#> 6: D2 2006-09-01 2025-08-11C. Data split
Function splitDataByDate extracts data by a specific
clinical event (e.g., first diagnosis dates of chronic diseases).
Users can define a table of clinical index dates of each patient. The
date can generate by selectCases function or first/last
admission date by getEligiblePeriod function.
This function can split data through classifying the data recorded before or after the defined index date and calculating the period between the record date and index date based on a self-defined window gap.
indexDateTable <- data.frame(ID = c("A0","B0","C0","D0"),
indexDate = c("2009-07-25", "2015-12-26",
"2015-12-05", "2017-01-29"),
stringsAsFactors = FALSE)
Data <- splitDataByDate(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
indexDateFile = indexDateTable,
gap = 30)
head(Data, 5)
#> ID ICD Date indexDate timeTag window
#> 1: A0 5856 2009-07-25 2009-07-25 A 1
#> 2: A0 V560 2010-03-28 2009-07-25 A 9
#> 3: A0 5853 2010-10-29 2009-07-25 A 16
#> 4: A0 V4511 2012-04-05 2009-07-25 A 33
#> 5: A0 5855 2013-12-20 2009-07-25 A 54D. Condition era
The concept of condition era is committed to the length of the persistence gap: when the time interval of any two consecutive admissions for certain conditions is smaller than the length of the persistence gap, then these two admission events will be aggregated into the same condition era.
Function getConditionEra calculates condition era by
using the grouped categories or self-defining groups of each patient and
then generates a table with individual IDs, the first and last record of
an era, and the sequence number of each episode.
Era <- getConditionEra(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015-10-01",
gapDate = 30,
groupDataType = ccs,
isDescription = FALSE)
head(Era)
#> ID CCS_CATEGORY era firstCaseDate endCaseDate count period
#> 1: A0 158 1 2009-07-25 2009-07-25 1 0 days
#> 2: A0 158 2 2010-03-28 2010-03-28 1 0 days
#> 3: A0 158 3 2010-10-29 2010-10-29 1 0 days
#> 4: A0 158 4 2012-04-05 2012-04-05 1 0 days
#> 5: A0 158 5 2013-12-20 2013-12-20 1 0 days
#> 6: A1 158 1 2006-11-29 2006-11-29 1 0 daysE. EDA preparation
After data integration, dxpr package provides a function to convert long format of grouped data into wide format which is fit to other analytical and plotting packages.
There are two type of output: numeric and binary
(numericOrBinary = B or N )
#binary
groupedData_Wide <- groupedDataLongToWide(dxDataFile = ELIX$groupedDT,
idColName = ID,
categoryColName = Comorbidity,
dateColName = Date,
numericOrBinary = B)
head(groupedData_Wide, 5)
#> ID ARTH CHRNLUNG DMCX DRUG HRENWORF HTNPREG LIVER LYMPH NEURO OBESE
#> 1: A0 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> 2: A1 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> 3: A10 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> 4: A11 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> 5: A12 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> PARA PERIVASC PSYCH RENLFAIL TUMOR ULCER VALVE WGHTLOSS
#> 1: FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
#> 2: FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
#> 3: FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
#> 4: FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
#> 5: FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
# numeric
groupedData_Wide <- groupedDataLongToWide(dxDataFile = ELIX$groupedDT,
idColName = ID,
categoryColName = Comorbidity,
dateColName = Date,
numericOrBinary = N)
head(groupedData_Wide, 5)
#> ID ARTH CHRNLUNG DMCX DRUG HRENWORF HTNPREG LIVER LYMPH NEURO OBESE PARA
#> 1: A0 0 0 0 0 0 0 0 0 0 0 0
#> 2: A1 0 0 0 0 0 0 0 0 0 0 0
#> 3: A10 0 0 0 0 0 0 0 0 0 0 0
#> 4: A11 0 0 0 0 0 0 0 0 0 0 0
#> 5: A12 0 0 0 0 0 0 0 0 0 0 0
#> PERIVASC PSYCH RENLFAIL TUMOR ULCER VALVE WGHTLOSS
#> 1: 0 0 5 0 0 0 0
#> 2: 0 0 5 0 0 0 0
#> 3: 0 0 5 0 0 0 0
#> 4: 0 0 5 0 0 0 0
#> 5: 0 0 5 0 0 0 0III. Visualization
Visualization provides overview of clinical data.
A. Pareto chart of error ICD list
Through code standardization, the functions
icdDxDecimalToShort and icdDxShortToDecimal
generate a table of diagnosis codes with potential errors.
The Pareto chart includes bar plot and line chart to visualize individual possible error ICD codes represented in descending order and cumulative total.
error <- icdDxDecimalToShort(dxDataFile = sampleDxFile,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015/10/01")
Plot_error1 <- plotICDError(errorFile = error$Error,
icdVersion = all,
wrongICDType = all,
others = TRUE,
topN = 10)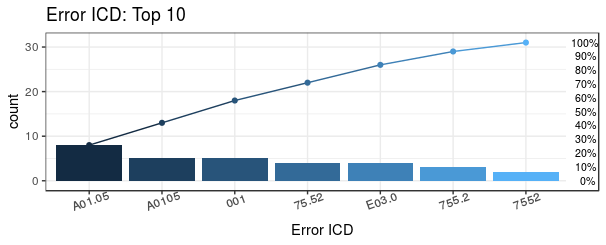
For instance, if a user chooses top 10 of common error ICD in dataset
(topN = 10), then the Pareto chart output
shows with top 10 error codes in this dataset and a list of the detail
of error ICD codes.
Plot_error1$ICD
#> ICD count CumCountPerc IcdVersionInFile WrongType Suggestion
#> 1: A0.11 20 18.35% ICD 10 Wrong format
#> 2: V27.0 18 34.86% ICD 10 Wrong version
#> 3: E114 8 42.2% ICD 10 Wrong format
#> 4: A01.05 8 49.54% ICD 9 Wrong version
#> 5: 42761 7 55.96% ICD 10 Wrong version
#> 6: Z9.90 6 61.47% ICD 10 Wrong format
#> 7: F42 6 66.97% ICD 10 Wrong format
#> 8: V24.1 6 72.48% ICD 10 Wrong version
#> 9: A0105 5 77.06% ICD 9 Wrong version
#> 10: 001 5 81.65% ICD 9 Wrong format 0019
#> 11: others 20 100% ICD 9 Wrong formatThe most common error ICD is A0.11 which has 20 admission records and error type is “wrong format”
Also, users can divide ICD-9 by the prefix of the
ICD code: 0, 1, 2,…, 9, V and E (groupICD = TRUE)
ICD-9-CM divided into 19 chapters:
001-139: Infectious And Parasitic Diseases
140-239: Neoplasms
….
For instance, if user chooses top 3 of common error ICD-9, the output Pareto chart shows with top 3 error codes and a list of the detail of error ICD codes.
Plot_error2 <- plotICDError(errorFile = error$Error,
icdVersion = 9,
wrongICDType = all,
groupICD = TRUE,
others = TRUE,
topN = 3)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> =
#> "none")` instead.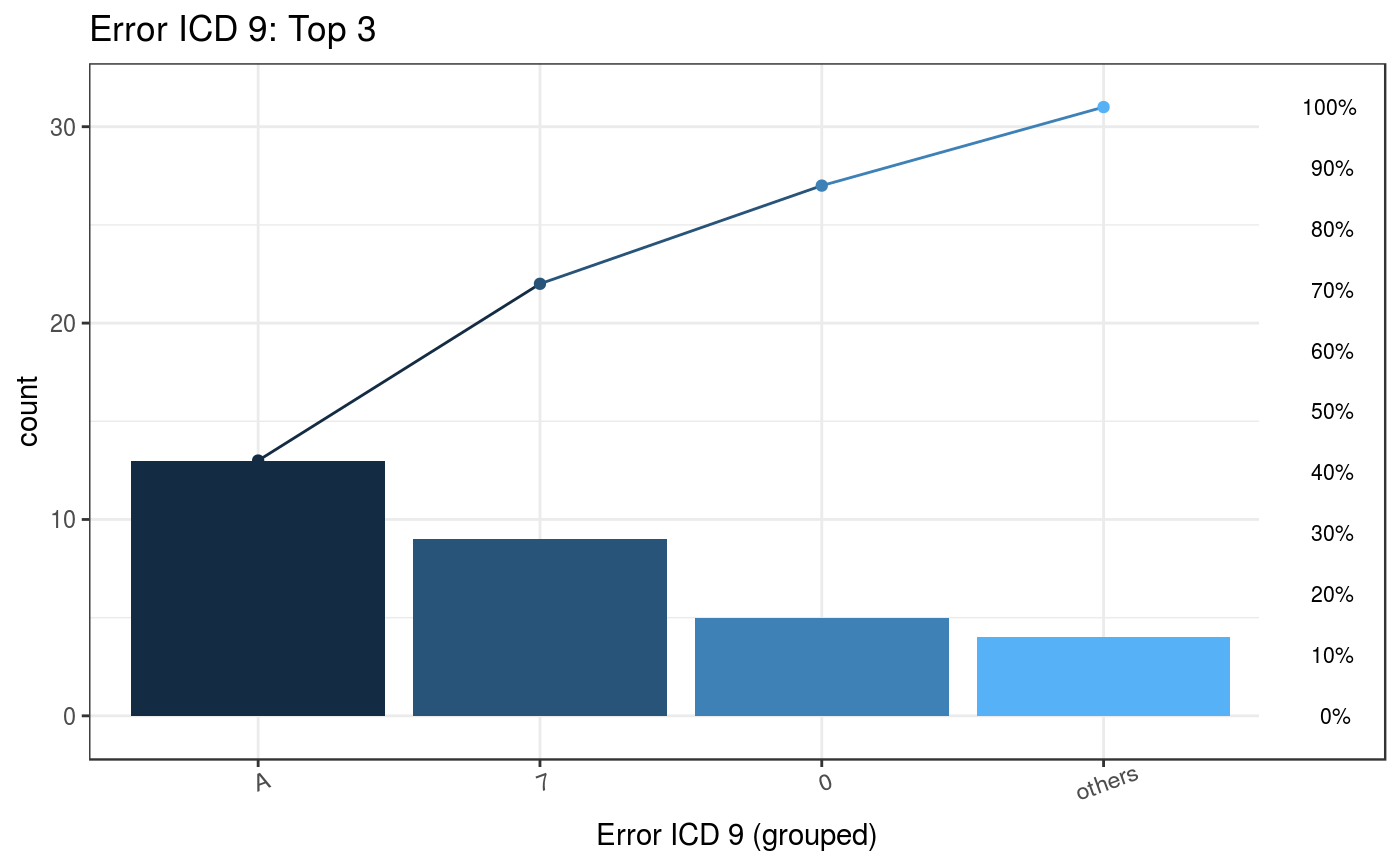
Plot_error2$ICD
#> ICDGroup groupCount CumCountPerc MostICDInGroup ICDPercInGroup WrongType
#> 1: A 13 41.94% A01.05 61.54% Wrong version
#> 2: 7 9 70.97% 75.52 44.44% Wrong format
00#> 3: 0 5 87.1% 001 100% Wrong format
#> 4: Others 4 100% E03.0 100% Wrong versionThe most common error ICD is A01.05 which has 13 admission records and the error type is “wrong version”.
B. histogram plot
plotDiagCat function provides an overview of grouping
category of the diagnostic code in histogram plot. User can observe the
proportion of diagnostic categories in their dataset.
groupedDataWide <- groupedDataLongToWide(ELIX$groupedDT,
idColName = ID,
categoryColName = Comorbidity,
dateColName = Date)
plot1 <- plotDiagCat(groupedDataWide = groupedDataWide,
idColName = ID,
topN = 10,
limitFreq = 0.01)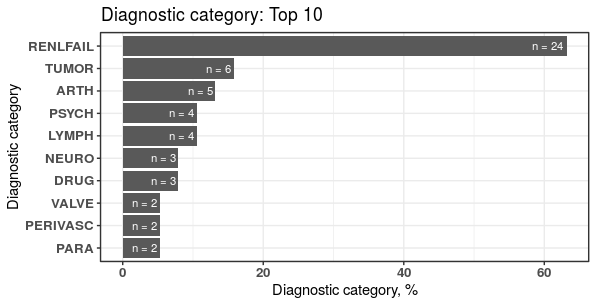
The first group, for instance, is grouped into “RENLFAIL” of ELIX comorbidity index.
plot1$sigCate
#> DiagnosticCategory N Percentage
#> 1: RENLFAIL 24 63.16
#> 2: TUMOR 6 15.79
#> 3: ARTH 5 13.16
#> 4: LYMPH 4 10.53
#> 5: PSYCH 4 10.53
#> 6: DRUG 3 7.89
#> 7: NEURO 3 7.89
#> 8: PARA 2 5.26
#> 9: PERIVASC 2 5.26
#> 10: VALVE 2 5.26This function can also do the Chi-square test and Fisher’s exact test to see if it is statistical significantly different between the diagnostic categories of case and control. The default level of significance is of 5% (p = 0.05).
selectedCaseFile <- selectCases(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015/10/01",
groupDataType = ccslvl2,
caseCondition = "Diseases of the urinary system",
caseCount = 1)
groupedDataWide <- groupedDataLongToWide(ELIX$groupedDT, ID, Comorbidity, Date,
selectedCaseFile = selectedCaseFile)
plot2 <- plotDiagCat(groupedDataWide = groupedDataWide,
idColName = ID,
groupColName = selectedCase,
topN = 10,
limitFreq = 0.01,
pvalue = 0.05)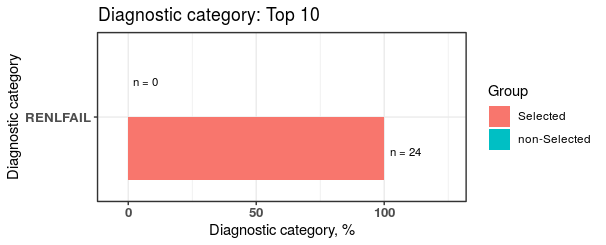
There are stastitcal significant difference in “RENLFAIL” of ELIX comorbidity index between case and control.
plot2$sigCate
#> DiagnosticCategory Group N Percentage
#> 1: RENLFAIL non-Selected 0 0
#> 2: RENLFAIL Selected 24 100Reference
I. Code transformation
ICD-9-CM code (2014): https://www.cms.gov/Medicare/Coding/ICD9ProviderDiagnosticCodes/codes.html
ICD-10-CM code (2019-2022): https://www.cms.gov/Medicare/Coding/ICD10
II. Code grouping
CCS (Clinical Classifications Software)
ICD-9-CM (2015): https://www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp
https://www.hcup-us.ahrq.gov/toolssoftware/ccs/Multi_Level_CCS_2015.zip
ICD-10-CM (2019): https://www.hcup-us.ahrq.gov/toolssoftware/ccsr/ccsr_archive.jsp
https://www.hcup-us.ahrq.gov/toolssoftware/ccs10/ccs_dx_icd10cm_2019_1.zip
CCSR (Clinical Classifications Software Refined)
ICD-10-CM (v2022-1): https://www.hcup-us.ahrq.gov/toolssoftware/ccsr/ccs_refined.jsp
PheWAS
ICD-9-Phecode (version 1.2, 2015): https://phewascatalog.org/phecodes
ICD-10 Phecode (version 1.2 beta, 2019): https://phewascatalog.org/phecodes_icd10cm
Comorbidities
ICD-9-AHRQ (2012-2015): https://www.hcup-us.ahrq.gov/toolssoftware/comorbidity/comorbidity.jsp#references
ICD-10-AHRQ (2019): https://www.hcup-us.ahrq.gov/toolssoftware/comorbidityicd10/comformat_icd10cm_2019_1.txt
ICD-9-Charlson: http://mchp-appserv.cpe.umanitoba.ca/Upload/SAS/ICD9_E_Charlson.sas.txt
ICD-10-Charlson: http://mchp-appserv.cpe.umanitoba.ca/Upload/SAS/ICD10_Charlson.sas.txt
ICD-9-Elixhauser (2012-2015): https://www.hcup-us.ahrq.gov/toolssoftware/comorbidity/comorbidity.jsp#references
ICD-10-Elixhauser (2019): https://www.hcup-us.ahrq.gov/toolssoftware/comorbidityicd10/comformat_icd10cm_2019_1.txt