5. NumPy
NumPy
- Basic NumPy
- Universal Functions (UFuncs) in NumPy
- Aggregations
- Broadcasting
- Boolean Arrays and Masks
NumPy
- Basic NumPy
- Universal Functions (UFuncs) in NumPy
- Aggregations
- Broadcasting
- Boolean Arrays and Masks
Basic NumPy
What is NumPy?
- Numerical Python
- Contain values of a single type
- the fundamental package for numerical computation in Python
- high-performance multidimensional array object + tools for working with these arrays
- foundation of other data science libraries (
Pandas,SciPy, andscikit-learn) - Key feature: ndarray n-dimensional array object
Why Use NumPy Arrays?
- Efficiency: more compact than Python lists
- Speed: vectorized operations on NumPy arrays are much faster than iterating through Python
lists(optimized C) - Functionality: provides a large library of mathematical functions
NumPy Getting Started
Install
Import
Create NumPy array (Recap)
Check Ch.3 for more information
np.full(dim, value): repeatedvalues- 2d dimension: (row, column)
Basic NumPy Array Manipulations
- Attributes: Size, shape, memory consumption, and data types of arrays
- Indexing: Getting and setting the value of individual array elements
- Slicing: Getting and setting smaller subarrays within a larger array
- Reshaping: Changing the shape of a given array
- Joining and splitting: Combining multiple arrays into one, and splitting one array into many
NumPy Array Attributes
- Every NumPy array has attributes:
ndim: the number of dimensionsshape: the size of each dimensionsize: the total size of the arraydtype: the data type of the array elementsitemsize: the size (in bytes) of each array elementnbytes: the total size (in bytes) of the array
NumPy Array Attributes - Examples
print(x2)
print("ndim: ", x2.ndim)
print("shape:", x2.shape)
print("size: ", x2.size)
print("dtype: ", x2.dtype)
print("itemsize:", x2.itemsize, "bytes")
print("nbytes: ", x2.nbytes, "bytes")[[3 5 2 4]
[7 6 8 8]
[1 6 7 7]]
ndim: 2
shape: (3, 4)
size: 12
dtype: int64
itemsize: 8 bytes
nbytes: 96 bytesNumPy Array Attributes - data type
- NumPy arrays contain values of a single type
dtype: Assign data type when creating a numpy array- Most of the time, this can be automatically detected
- Available data type: ref
Array Indexing - Single Elements
- Similar to lists
- In a multi-dimensional array, items can be accessed using a comma-separated indices
- Indexing starts from 0
Array Indexing - Example
x[row, column]-1: last index
Array Slicing - Subarrays
- Colon (
:) operator: slice arrays x[start:stop:step]- Default:
start=0stop=size of dimensionstep=1
Array Slicing - Example (1D)
x[start:stop:step]
Array Slicing - Example (2D)
No Copy View
- In NumPy array, slices will be View. in lists, slices will be copies
.copy()if you need a copy
Reshaping of Arrays
reshape(dim): gives a new shape to an array- the size must match
Hands-on - Indexing and Slicing - 1
You’re working on a project to analyze and manipulate digital images using NumPy. Your task is to perform various operations on image data represented as NumPy arrays.
import numpy as np
import matplotlib.pyplot as plt
# Create a sample 8x8 grayscale image (0-255 values)
image = np.array([
[50, 50, 50, 50, 200, 200, 200, 200],
[50, 50, 50, 50, 200, 200, 200, 200],
[50, 90, 90, 50, 200, 200, 200, 200],
[50, 90, 90, 50, 200, 200, 200, 200],
[50, 50, 50, 50, 200, 200, 200, 200],
[50, 50, 50, 50, 200, 200, 200, 200],
[50, 50, 50, 50, 50, 50, 50, 50],
[50, 50, 50, 50, 50, 50, 50, 50]
])Hands-on - Indexing and Slicing - 2
Hands-on - Indexing and Slicing - 3
- Extract the 4x4 sub-image from the top-left corner.
- Extract every other pixel from both dimensions (creating a 4x4 image).
- Flatten the image into a 1D array.
- Reshape the flattened array back into an 8x8 image.
Array Concatenation and Splitting
np.concatenate([array1, array2, ...]): Concatenates arrays along an existing (or first) axis.np.vstack([array1, array2]): Vertical stack (row-wise concatenation).np.hstack([array1, array2]): Horizontal stack (column-wise concatenation).
Concatenation - Example
np.concatenate([array1, array2, ...]): Concatenates arrays along an existing (or first) axis.
Concatenation - Example
np.concatenate([array1, array2, ...]): Concatenates arrays along an existing (or first) axis.
array([[1, 2, 3],
[4, 5, 6],
[1, 2, 3],
[4, 5, 6]])Set axis=1 for the second axis
Concatenation - np.vstack
np.vstack([array1, array2]): Vertical stack (row-wise concatenation).
Concatenation - np.hstack
np.hstack([array1, array2]): Horizontal stack (column-wise concatenation).
Splitting
The opposite of concatenation is splitting
np.split(np array,[split points])ornp.split(np array, # of sections)np.vsplit(np array,[split points])ornp.vsplit(np array, # of sections)np.hsplit(np array,[split points])ornp.hsplit(np array, # of sections)- split points: split before the split point
- N split-points, leads to N + 1 subarrays
- # of sections: divided into N equal arrays
Splitting
split points
[1 2 3] [99 99] [3 2 1]number of sections
Splitting - np.vsplit
[[1 2 3]
[3 2 1]
[1 2 3]]np.vsplit(np array,[split points]) or np.vsplit(np array, # of sections)
Splitting - np.hsplit
[[1 2 3]
[3 2 1]
[1 2 3]]np.hsplit(np array,[split points]) or np.hsplit(np array, # of sections)
Hands-on - Concatenation & Split - 1
You’re a meteorologist working on analyzing and combining weather data from multiple stations.
import numpy as np
import matplotlib.pyplot as plt
# Generate sample weather data for 3 stations over 5 days
np.random.seed(42)
station1 = np.random.randint(15, 25, size=(5, 3)) # 5 days, temp/humidity/wind
station2 = np.random.randint(18, 28, size=(5, 3))
station3 = np.random.randint(20, 30, size=(5, 3))
print("Station 1 data:")
print(station1)Station 1 data:
[[21 18 22]
[19 21 24]
[17 21 22]
[19 18 22]
[22 17 20]]Hands-on - Concatenation & Split - 2
- Combine the data from all three stations vertically (stacking days).
- Combine the data from all three stations horizontally (side by side).
- Split the vertically stacked data back into three station datasets.
- Split the horizontally stacked data into temperature, humidity, and wind speed datasets.
NumPy
- Basic NumPy
- Universal Functions (UFuncs) in NumPy
- Aggregations
- Broadcasting
- Boolean Arrays and Masks
Universal Functions (UFuncs) in NumPy
Why UFuncs?
- The key to vectorized operations in NumPy.
- Allow you to apply a function element-wise to arrays
- with significant performance gains compared to Python loops.
The Slowness of Loops
- Python’s default implementation can be slow for repeated small operations.
- Dynamic typing and interpreted nature lead to overhead in loops.
UFuncs to the Rescue
- Fast, vectorized alternative to loops
big_array = np.random.randint(1, 100, size=1000000)
#Compare the time between looping and ufuncs.
%timeit compute_reciprocals(big_array) #previous slide's method
%timeit (1.0 / big_array) #ufunc implementation1.27 s ± 8.39 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
1.94 ms ± 17.4 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)UFuncs: Two Flavors
- Unary UFuncs: Operate on a single input array. Examples:
np.abs,np.sin,np.exp. - Binary UFuncs: Operate on two input arrays. Examples:
np.add,np.subtract,np.multiply,np.divide,np.power.
Array Arithmetic
x = np.arange(4)
print("x =", x)
print("x + 5 =", x + 5) #np.add(x,5)
print("x - 5 =", x - 5) #np.subtract(x,5)
print("x * 2 =", x * 2) #np.multiply(x,2)
print("x / 2 =", x / 2) #np.divide(x,2)
print("x // 2 =", x // 2) # Floor division
print("-x =", -x)
print("x ** 2 =", x ** 2)
print("x % 2 =", x % 2)x = [0 1 2 3]
x + 5 = [5 6 7 8]
x - 5 = [-5 -4 -3 -2]
x * 2 = [0 2 4 6]
x / 2 = [0. 0.5 1. 1.5]
x // 2 = [0 0 1 1]
-x = [ 0 -1 -2 -3]
x ** 2 = [0 1 4 9]
x % 2 = [0 1 0 1]UFunc Equivalents
| Operator | Equivalent UFunc | Description |
|---|---|---|
+ |
np.add |
Addition |
- |
np.subtract |
Subtraction |
- |
np.negative |
Unary negation |
* |
np.multiply |
Multiplication |
/ |
np.divide |
Division |
// |
np.floor_divide |
Floor division |
** |
np.power |
Exponentiation |
% |
np.mod |
Modulus/remainder |
Absolute Value
Trigonometric Functions
- Trigonometric functions in NumPy
np.sin(),np.cos(),np.tan()
# Return evenly spaced numbers over a specified interval
theta = np.linspace(0, np.pi, 3) #
print("theta =", theta)
print("sin(theta) =", np.sin(theta))
print("cos(theta) =", np.cos(theta))
print("tan(theta) =", np.tan(theta))theta = [0. 1.57079633 3.14159265]
sin(theta) = [0.0000000e+00 1.0000000e+00 1.2246468e-16]
cos(theta) = [ 1.000000e+00 6.123234e-17 -1.000000e+00]
tan(theta) = [ 0.00000000e+00 1.63312394e+16 -1.22464680e-16]Trigonometric Functions
- Inverse trigonometric functions are also available
np.arcsin(),np.arccos(),np.arctan()
Exponents
np.exp(), np.exp2(), np.power()
Logarithms
np.log()(natural log)np.log2()(base-2)np.log10()(base-10)
Hands-on - UFuncs - 1/2
You’re analyzing sensor data from industrial equipment. The dataset contains temperature (℃), vibration (mm/s²), and pressure (kPa) readings sampled every 5 minutes over 30 days.
# Generate synthetic sensor data (4320 = 30 days * 144 samples/day)
np.random.seed(42)
time_points = 4320
temperature = 25 + 10 * np.sin(2 * np.pi * np.arange(time_points)/144) + np.random.normal(0, 1, time_points)
vibration = np.abs(2 * np.random.randn(time_points) + np.sin(np.arange(time_points)/100))
pressure = 100 + 20 * np.cos(2 * np.pi * np.arange(time_points)/288) + np.random.normal(0, 3, time_points)Hands-on - UFuncs - 2/2
- Remove negative vibration values using
np.clip - Find daily temperature amplitude (max - min)
- Calculate equipment stress using custom formula with UFuncs
- stress = (temp² + vib²) / pressure
- Apply log transform (natural log) to vibration data
Specialized UFuncs (FYR)
- NumPy has many more ufuncs
scipy.specialis an excellent source for more specialized mathematical functions.- Examples from
scipy.specialgamma(),gammaln(),beta(),erf(),erfc(),erfinv()
Specialized UFuncs (FYR) - Examples
from scipy import special
import numpy as np
# Gamma functions and related functions
x = [1, 5, 10]
print("gamma(x) =", special.gamma(x))
print("ln|gamma(x)| =", special.gammaln(x))
print("beta(x, 2) =", special.beta(x, 2))gamma(x) = [1.0000e+00 2.4000e+01 3.6288e+05]
ln|gamma(x)| = [ 0. 3.17805383 12.80182748]
beta(x, 2) = [0.5 0.03333333 0.00909091]NumPy
- Basic NumPy
- Universal Functions (UFuncs) in NumPy
- Aggregations
- Broadcasting
- Boolean Arrays and Masks
Aggregations
Why Aggregations?
- Often, the first step when facing large datasets is to compute summary statistics.
- Aggregations provide a concise way to describe the “typical” values in a dataset.
- Examples: mean, standard deviation, sum, product, min, max, median, quantiles.
Summing Array Values
- Python’s built-in
sum()function:
- NumPy’s
np.sum()function:
NumPy’s np.sum() is Faster
Minimum and Maximum
- Python’s built-in
min()andmax()functions
(3.7745761005680833e-07, 0.9999993822414859)- NumPy’s
np.min()andnp.max()functions
NumPy’s np.min() and np.max() are Faster!
Shorter Syntax: Array Methods
- For
min,max,sum, etc., use the array object’s methods:
Multi-Dimensional Aggregates
- Aggregate along rows or columns in multi-dimensional arrays using the
axisargument. - Default: across all values
axis Argument Example (Columns)
axis=0: collapses the first axis
axis Argument Example (Rows)
axis=1: collapses the second axis
Other Aggregation Functions - 1
np.sum(np.nansum): Compute sum of elementsnp.prod(np.nanprod): Compute product of elementsnp.mean(np.nanmean): Compute mean of elementsnp.std(np.nanstd): Compute standard deviationnp.var(np.nanvar): Compute variancenp.min(np.nanmin): Find minimum valuenp.max(np.nanmax): Find maximum value
Other Aggregation Functions - 2
np.argmin(np.nanargmin): index of minimum valuenp.argmax(np.nanargmax): index of maximum valuenp.median(np.nanmedian): median of elementsnp.percentile(np.nanpercentile): rank-based statistics of elementsnp.any: whether any elements are truenp.all: whether all elements are true
Example: 前置作業
為了成功從https (加密封包傳輸)下載資料,首先取消證書驗證
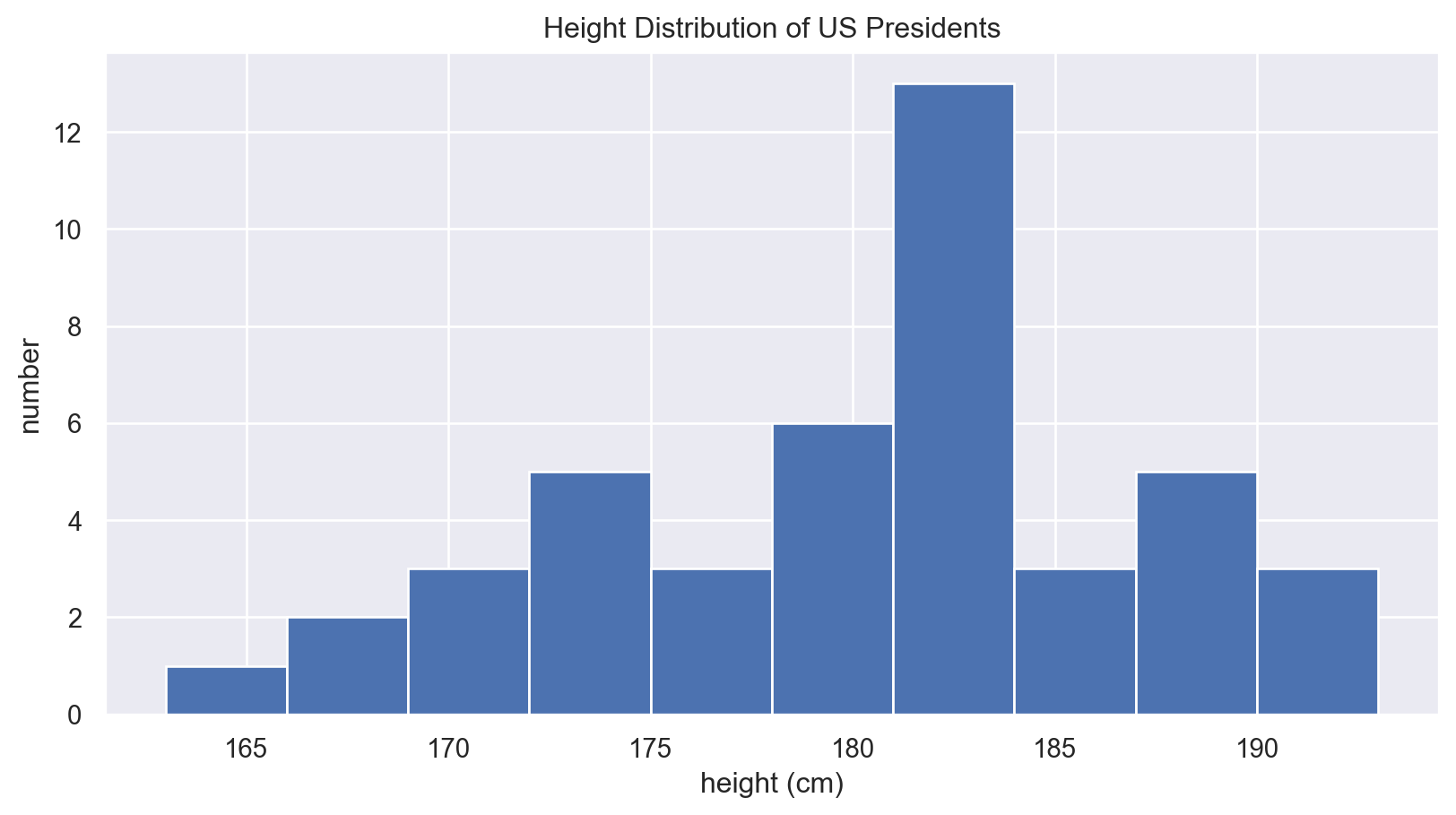
!pip3 install seabornExample: President Heights
- Uses a real-world dataset to demonstrate aggregates.
import numpy as np
import pandas as pd
data = pd.read_csv('https://raw.githubusercontent.com/jakevdp/PythonDataScienceHandbook/refs/heads/master/notebooks/data/president_heights.csv')
heights = np.array(data['height(cm)'])
print("Mean height: ", heights.mean())
print("Standard deviation:", heights.std())
print("Minimum height: ", heights.min())
print("Maximum height: ", heights.max())
print("25th percentile: ", np.percentile(heights, 25))
print("Median: ", np.median(heights))
print("75th percentile: ", np.percentile(heights, 75))Mean height: 180.04545454545453
Standard deviation: 6.983599441335736
Minimum height: 163
Maximum height: 193
25th percentile: 174.75
Median: 182.0
75th percentile: 183.5Visualizing President Heights
Hands-on - Aggregations - 1/2
You’re a financial analyst tasked with analyzing historical stock data for several tech companies.
np.random.seed(42)
companies = ['TechCorp', 'DataSys', 'AIGlobal', 'CloudNet', 'CyberSec']
trading_days = 252
stock_data = np.random.randint(100, 200, size=(len(companies), trading_days))
stock_data = stock_data * (1 + np.random.randn(len(companies), trading_days) * 0.01) # Add some randomness
print("Stock Data Shape:", stock_data.shape)
print("First six days of data:\n", stock_data[:, :6])Stock Data Shape: (5, 252)
First six days of data:
[[149.4074983 190.59940972 112.19491634 173.42047559 158.72559527
120.79293028]
[196.30842854 174.9778714 157.40229724 169.92405711 178.52343482
191.48421877]
[186.46131515 155.38937619 126.11957816 175.40848392 188.43959037
169.95625021]
[190.52887619 106.23981577 156.97119204 161.0857364 147.58640448
124.25445626]
[123.82077119 161.16804061 194.75072474 158.84621237 155.07543654
159.51183387]]Hands-on - Aggregations - 2/2
- Calculate for each company:
- Average stock price
- Highest and lowest stock prices
- Standard deviation of stock prices
- Number of days the stock price increased
- Hint:
np.diff
- Hint:
- Get the day with the highest average stock price across all companies
- Get the company with the most volatile stock (highest standard deviation)
NumPy
- Basic NumPy
- Universal Functions (UFuncs) in NumPy
- Aggregations
- Broadcasting
- Boolean Arrays and Masks
Broadcasting
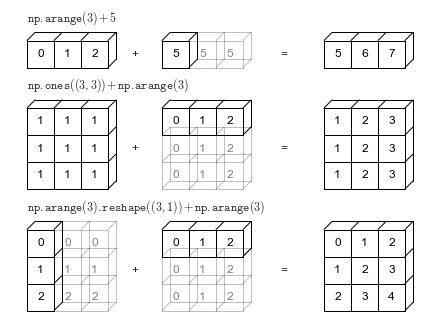
Broadcasting: The Basic Idea
Enabling UFuncs to operate on arrays of different sizes
- Binary operations on arrays of the same size operate element-wise.
- Broadcasting allows operations on arrays of different sizes.
Broadcasting Analogy
“stretching” or “duplicating” the smaller array to match the shape of the larger array
Broadcasting Example: Higher Dimensions
The 1D array a is “broadcast” across the 2nd dimension of M
Broadcasting Example: Both Arrays
Both a and b are broadcast to a common shape
Rules of Broadcasting
- Rule 1: If the two arrays differ in their number of dimensions, the shape of the one with fewer dimensions is padded with ones on its leading (left) side.
- Rule 2: If the shape of the two arrays does not match in any dimension, the array with shape equal to 1 in that dimension is stretched to match the other shape.
- Rule 3: If in any dimension the sizes disagree and neither is equal to 1, an error is raised.
Broadcasting Example 1
[[1. 1. 1.]
[1. 1. 1.]]
[0 1 2]
[[1. 2. 3.]
[1. 2. 3.]]- Shapes:
M.shape = (2, 3)a.shape = (3)
- Rule 1: Pad
awith ones:M.shape -> (2, 3)a.shape -> (1, 3)
- Rule 2: Stretch the first dimension of
a:M.shape -> (2, 3)a.shape -> (2, 3)
- Result: Shapes match!
Broadcasting Example 2
[[0]
[1]
[2]]
[0 1 2]
[[0 1 2]
[1 2 3]
[2 3 4]]- Shapes:
a.shape = (3, 1)b.shape = (3)
- Rule 1: Pad
bwith ones (left):a.shape -> (3, 1)b.shape -> (1, 3)
- Rule 2: Stretch both arrays:
a.shape -> (3, 3)b.shape -> (3, 3)
- Result: Shapes match!
Broadcasting Example 3: Incompatible
M = np.ones((3, 2))
a = np.arange(3)
print(M)
print(a)
# This will raise a ValueError:
# print(M + a)[[1. 1.]
[1. 1.]
[1. 1.]]
[0 1 2]- Shapes:
M.shape = (3, 2)a.shape = (3,)
- 1: Pad
awith ones:M.shape -> (3, 2)a.shape -> (1, 3)
- 2: Stretch the first dimension of
a:M.shape -> (3, 2)a.shape -> (3, 3)
- 3: Shapes disagree, and neither dimension is 1. Error!
Broadcasting in Practice - 1
Centering an Array
[[0.39543427 0.31320626 0.14706185]
[0.4878512 0.13004778 0.83548024]
[0.37039699 0.37480357 0.47400643]
[0.44412771 0.99485797 0.54538112]
[0.0882017 0.60687734 0.15158429]
[0.03093764 0.97074215 0.77353889]
[0.98010899 0.47787845 0.53225753]
[0.16833696 0.24690743 0.83854921]
[0.35618565 0.82681497 0.70901372]
[0.17717093 0.50744637 0.67738732]]Broadcasting in Practice - 2
[ 4.99600361e-17 -9.99200722e-17 -6.66133815e-17]Broadcasting allows subtracting the feature means from each observation efficiently.
Hands-on - Broadcasting
You have temperature data for multiple cities over a week.
temperatures = np.random.randint(15, 35, size=(5, 7))
cities = ['New York', 'Los Angeles', 'Chicago', 'Houston', 'Phoenix']
days = ['Mon', 'Tue', 'Wed', 'Thu', 'Fri', 'Sat', 'Sun']
print(temperatures)[[26 25 21 18 33 15 17]
[27 29 16 25 18 33 31]
[17 32 22 33 19 21 22]
[19 31 19 18 33 16 25]
[27 29 26 25 21 19 31]]- Convert all temperatures from Celsius to Fahrenheit.
- Rank the temperatures for each day across cities (1 being the hottest, 5 being the coldest).
np.argsort() - Calculate a 3-day weighted moving average for each city, where the weights are [0.6, 0.3, 0.1] for [today, yesterday, day before yesterday].
np.dot()
NumPy
- Basic NumPy
- Universal Functions (UFuncs) in NumPy
- Aggregations
- Broadcasting
- Boolean Arrays and Masks
Boolean Arrays and Masks
Comparison Operators
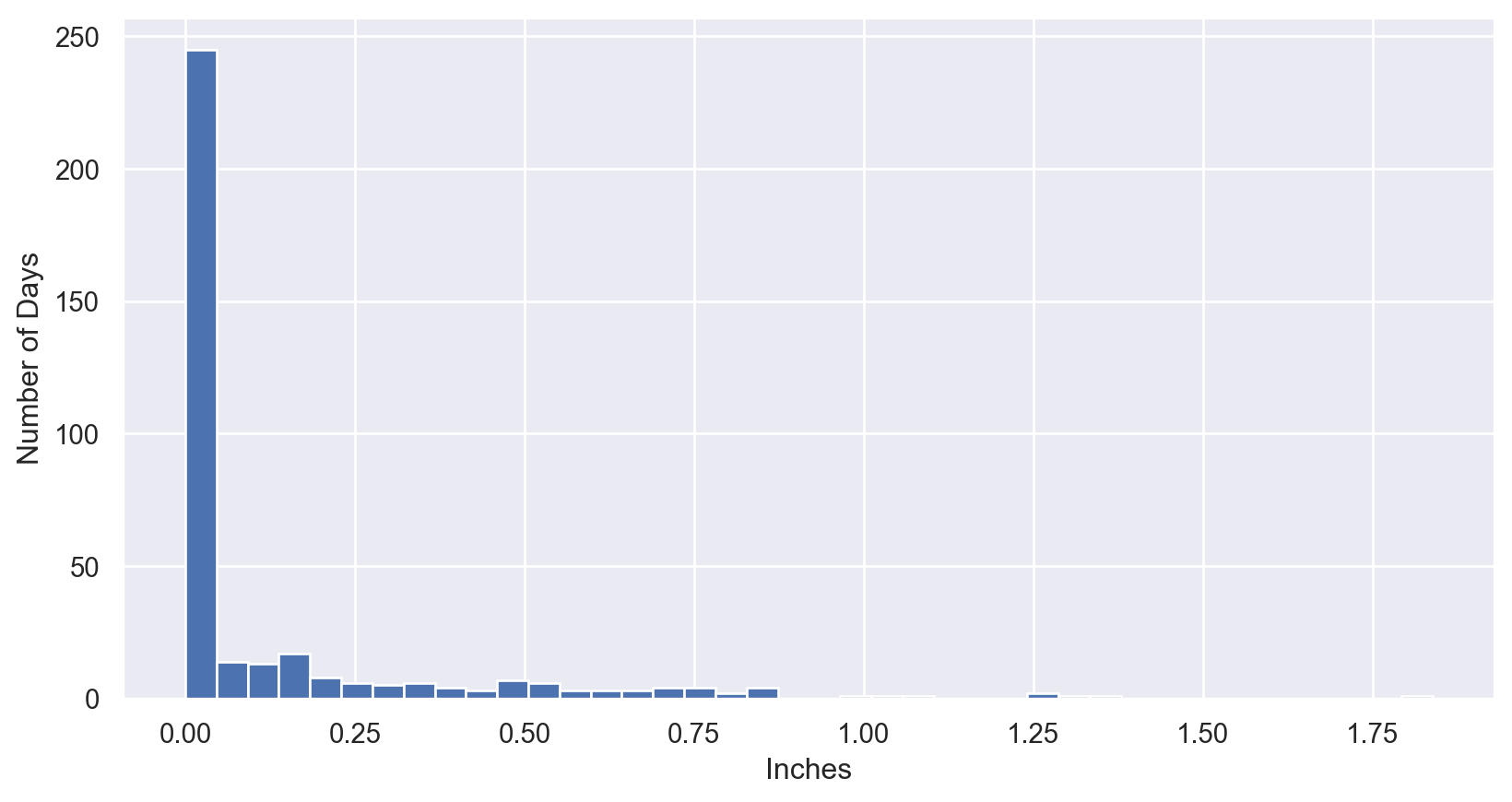
Rainfall Data: Create the Plot
Rainfall Data: Boolean Masking
rainfall = pd.read_csv('https://raw.githubusercontent.com/amankharwal/Website-data/refs/heads/master/Seattle2014.csv')['PRCP'].values
inches = rainfall / 254 # 1/10mm -> inches
print("Number days without rain: ", np.sum(inches == 0))
print("Number days with rain: ", np.sum(inches != 0))
print("Days with more than 0.5 inches:", np.sum(inches > 0.5))
print("Rainy days with < 0.2 inches :", np.sum((inches > 0) &
(inches < 0.2)))Number days without rain: 215
Number days with rain: 150
Days with more than 0.5 inches: 37
Rainy days with < 0.2 inches : 75any() and all()
any(): Are any values True?all(): Are all values True?- Can be used along axes, like aggregates.